Module 1.2
Getting started: managing data
Spring 2024
Load script for module #1.2
Click here to download the script! Save the script to the project directory you set up in the previous module.
Load your script in RStudio. To do this, open RStudio and click on the folder icon in the toolbar at the top to load your script.
Let’s get started with data management in R!
Working directory
The working directory is the first place R looks for any files you would like to read in (e.g., code, data). It is also the first place R will try to write any files you want to save.
It makes things a lot easier if you put all your data files into a single folder and tell R to make that folder your working directory. That way, you won’t need to wrangle complex directory names!
Every R session has a working directory, whether you specify one or not. Let’s find out what my working directory is right now:
# Working directories --------------------
# Find the directory you're working in
getwd() # note: the results from running this command on my machine will differ from yours! ## [1] "C:/Users/Kevin/Documents/GitHub/R-Bootcamp"What’s yours? Usually the default working directory is your “Documents” folder.
Since you should already be working in an RStudio Project, your working directory will be set as your project directory (directory that contains the .Rproj file for your project). This is convenient, because:
- That’s most likely where the data for that project live anyway
- If you’re collaborating with someone else on the project (e.g., in a shared Dropbox folder), you can both open the project and R will instantly know where to read and write data without either of you having to reset the working directory (even though the directory path is probably different on your two machines). This can save a lot of headaches!
Reminder: to start a new RStudio Project, just click on “File->New Project” in RStudio’s menu bar.
Set your working directory
You can set a new working directory using the “setwd()” function.
# for example...
# setwd("E:/GIT/R-Bootcamp") # note that the use of backslashes for file paths, as used by Windows, are not supported by RNOTE: when you put file paths in R, they need to use forward slashes (“/”; or double backslashes, “\\”) – single backslashes (“\”, as seen in Windows) do not work for specifying file paths in R.
Alternatively, you can (in RStudio) use the dropdown menus at the top to set the working directory (Session->Set Working Directory->Choose Directory).
Once you have set the working directory, you can use the “list.files()” function to see what’s in the directory. If I ran this, we would see the contents of the directory that I’m using to create this website!
# Contents of working directory
# list.files() # uncomment this to run it...What’s in your working directory?
Importing data into R!
There are many ways data can be imported into R. Many types of files can be imported (e.g., text files, csv, shapefiles). And people are always inventing new ways to read and write data to/from R. But here are the basics.
- read.table or read_delim
- reads a data file in any major text format (comma-delimited, space
delimited etc.), you can specify which format (very general)
- reads a data file in any major text format (comma-delimited, space
delimited etc.), you can specify which format (very general)
- read.csv or read_csv
- fields are separated by a comma (this is the most common way to read in data)
Before we can read data in, we need to put some data files in our working directory!
NOTE: you can also read data from Excel (.xlsx files) directly using the ‘readxl’ package.
Download the following data files and store them in your working directory (i.e., the folder where your scripts are already!)
Make sure the data files are stored in your working directory (project directory)!
Once you have saved these files to your working directory, open one or two of them up (e.g., in Excel or a text editor) to see what’s inside.
Now let’s read them into R!!
# Import/Export data files into R----------------------
# read_csv to import textfile with columns separated by commas
data.df <- read_csv("data.csv")
names(data.df)
# Remove redundant objects from your workspace
rm(data.txt.df)Using R’s built in data
R has many useful built-in data sets that come pre-loaded. You can explore these datasets with the following command:
# Built-in data files -----------------------
data() Let’s read in one of these datasets!
data(mtcars) # read built-in data on car road tests performed by Motor Trend
head(mtcars) # inspect the first few lines
# ?mtcars # learn more about this built-in data setMany packages come with built-in datasets as well. For, instance, ggplot2 comes with the “diamonds” package:
ggplot2::diamonds # note the use of the package name followed by two colons- this is a way to make sure you are using a function (or data set or other object) from a particular package... (sometimes several packages have functions with the same name...)## # A tibble: 53,940 × 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
## 4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
## # ℹ 53,930 more rowsBasic data checking
To learn more about the ‘internals’ of any data object in R, we can use the “str()” (structure) function:
# Check/explore data objects --------------------------
# ?str: displays the internal structure of the data object
str(mtcars)## 'data.frame': 32 obs. of 11 variables:
## $ mpg : num 21 21 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 ...
## $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
## $ disp: num 160 160 108 258 360 ...
## $ hp : num 110 110 93 110 175 105 245 62 95 123 ...
## $ drat: num 3.9 3.9 3.85 3.08 3.15 2.76 3.21 3.69 3.92 3.92 ...
## $ wt : num 2.62 2.88 2.32 3.21 3.44 ...
## $ qsec: num 16.5 17 18.6 19.4 17 ...
## $ vs : num 0 0 1 1 0 1 0 1 1 1 ...
## $ am : num 1 1 1 0 0 0 0 0 0 0 ...
## $ gear: num 4 4 4 3 3 3 3 4 4 4 ...
## $ carb: num 4 4 1 1 2 1 4 2 2 4 ...str(data.df)## spc_tbl_ [20 × 4] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
## $ Country: chr [1:20] "Bolivia" "Brazil" "Chile" "Colombia" ...
## $ Import : num [1:20] 46 74 89 77 84 89 68 70 60 55 ...
## $ Export : num [1:20] 0 0 16 16 21 15 14 6 13 9 ...
## $ Product: chr [1:20] "N" "N" "N" "A" ...
## - attr(*, "spec")=
## .. cols(
## .. Country = col_character(),
## .. Import = col_double(),
## .. Export = col_double(),
## .. Product = col_character()
## .. )
## - attr(*, "problems")=<externalptr>names(diamonds)## [1] "carat" "cut" "color" "clarity" "depth" "table" "price"
## [8] "x" "y" "z"And we can use the “summary()” function to get a brief summary of the contents of each column in our data frame:
summary(data.df)## Country Import Export Product
## Length:20 Min. :35.0 Min. : 0.00 Length:20
## Class :character 1st Qu.:66.0 1st Qu.: 3.00 Class :character
## Mode :character Median :74.0 Median : 8.00 Mode :character
## Mean :72.1 Mean : 9.55
## 3rd Qu.:84.0 3rd Qu.:15.25
## Max. :91.0 Max. :23.00Exporting and saving data in R
Reading in data is one thing, but you will probably also want to write data to your hard drive as well. There are countless reasons for this- you might want to use an external program to plot your data, you might want to archive some simulation results.
Again, there are many ways to write data to a file. Here are the basics!
Exporting data as table
# Exporting data (save to hard drive as data file)
# ?write_csv: writes a CSV file to the working directory
newdf <- data.df[,c("Country","Product")]
write_csv(newdf, file="data_export.csv") # export a subset of the data we just read in.Saving (and loading) R data objects (binary)
The data loaded in your R environment are stored in your computer’s memory as binary representations that are efficient but not human-readable (this is how computers store and manage data). But sometimes the lack of human readability isn’t a problem. For example: what if all we want to do is save the data so that we can load it back into an R session at a later date? In this case we never need to look at the data outside of R.
To save data we can use the “saveRDS()” and “save()” functions. To load data we can use the “readRDS()” and “load()” functions:
NOTE: binary R data files should be stored with the extensions “.RData”:
# Saving and loading
# saveRDS: save a single object from the environment to hard disk
# save: saves one or more objects from the environment to hard disk. Must be read back in with the same name.
a <- 1
b <- data.df$Product
saveRDS(b, "Myobject1.rds") # use saveRDS to save individual R objects
save(a,b,file="Myobjects1.RData") # use 'save' to save sets of objects
save.image("Myworkspace.RData") # use 'save.image' to save your entire workspace
rm(a,b) # remove these objects from the environment
new_b <- readRDS("Myobject1.rds")
load("Myworkspace.RData") # load these objects back in with the same names!Clearing the environment
Sometimes your environment can get cluttered with objects. In these cases, it can help to clear the environment. You can just use the ‘broom’ icon in your environment window in RStudio to clear your environment, or use this command:
# Clear the environment
rm(list=ls()) # clear the entire environment. Confirm that your environment is now empty!
data.df <- read_csv("data.csv") # read the data back in!Working with data in R
Now let’s start seeing what we can do with data in R. Even without doing any statistical analyses, R is very a powerful environment for doing data transformations and performing mathematical operations.
Boolean operations
Boolean operations refer to TRUE/FALSE tests. That is, we can ask a question about the data to a Boolean operator and the operator will return a TRUE or a FALSE (logical) result.
First, let’s meet the boolean operators.
NOTE: don’t get confused between the equals sign (“=”), which is an assignment operator (same as “<-”), and the double equals sign (“==”), which is a Boolean operator:
# Working with data in R ------------------------
# <- assignment operator
# = alternative assignment operator
a <- 3 # assign the value 3 to the object named "a"
b = 5 # assign the value 5 to the object named "b"
a == 3 # answer the question: "does the object "a" equal "3"? ## [1] TRUEa == b## [1] FALSE# what happens if you accidentally typed 'a = b'?# Boolean operations
# Basic operators
# < less than
# > greater than
# <= less than or equal to
# >= greater than or equal to
# == equal to
# != not equal to
# %in% belongs to a set
# Combining multiple conditions
# & must meet both conditions (AND operator)
# | must meet one of two conditions (OR operator)
# explore Boolean operators -----------------------
Y <- 4 # first define a couple new objects
Z <- 6
Y == Z # is Y equal to Z? (T/F)
Y < Z # is Y less than Z?
!(Y < Z) # the exclamation point reverses any boolean object (read "NOT")
data.df[,2]=74 # (for each element in the second column of data.df) is it equal to 74?
# OOPS! sets entire second column equal to 74! OOPS WE GOOFED UP!!!
data.df <- read.csv("data.csv") ## correct our mistake in the previous line (revert to the original data)!
# Let's do it right this time
data.df[,2]==74 # tests each element of column to see whether it is equal to 74
data.df[,2]<74|data.df[,2]==91 # combine two questions using the logical OR operatorThis image from Wickham and Grolemund’s R for Data Science book can help conceptualize the combination operators:
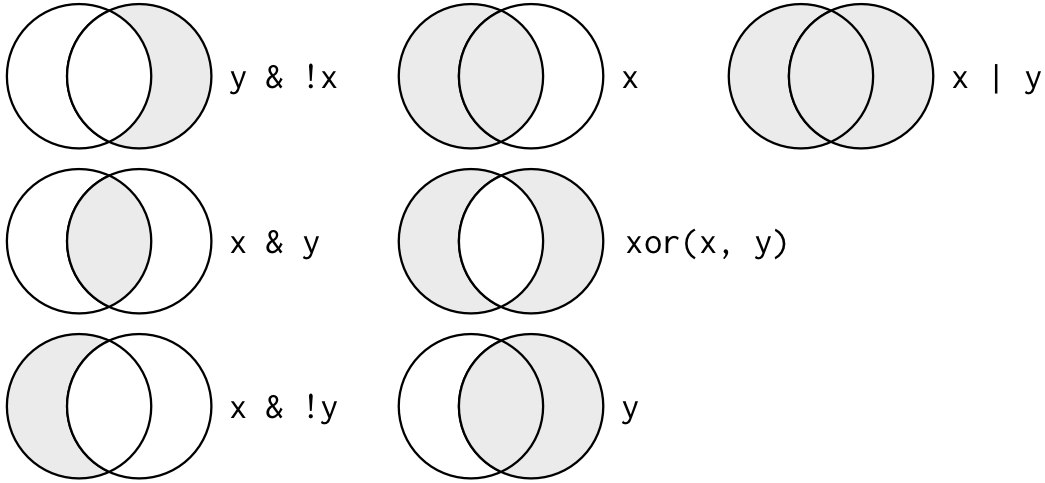
Data subsetting using boolean logic
Boolean operations are great for subsetting data - that is, we can select only those rows/observations that meet a certain condition (e.g., where [some condition] is TRUE).
# Subsetting data
data.df[data.df[,"Import"]<74,] # select those rows of data for which second column is less than 74## # A tibble: 9 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 DominicanRep 68 14 A
## 3 Ecuador 70 6 A
## 4 ElSalvador 60 13 A
## 5 Guatemala 55 9 A
## 6 Haiti 35 3 A
## 7 Honduras 51 7 A
## 8 Nicaragua 68 0 A
## 9 Peru 73 0 N# or alternatively, using tidyverse syntax (and the pipe operator):
data.df %>%
filter(Import<74) # use "filter" verb from 'dplyr' package## # A tibble: 9 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 DominicanRep 68 14 A
## 3 Ecuador 70 6 A
## 4 ElSalvador 60 13 A
## 5 Guatemala 55 9 A
## 6 Haiti 35 3 A
## 7 Honduras 51 7 A
## 8 Nicaragua 68 0 A
## 9 Peru 73 0 Nsub.countries<-c("Chile","Colombia","Mexico") # create vector of character strings
data.df %>%
filter(Country %in% sub.countries) # subset the dataset for only that subset of countries we're interested in## # A tibble: 3 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Chile 89 16 N
## 2 Colombia 77 16 A
## 3 Mexico 83 4 NPractice data processing!
Let’s switch to a different data file. Download the Turtle Data and save to your project directory.
Let’s use this data set to review the summarizing and subsetting operations we have learned- and learn a few new things along the way:
Subsetting data
# Subsetting data ----------------------
# Practice subsetting a data frame
turtles.df <- read_delim(file="turtle_data.txt",delim="\t") # tab-delimited file
turtles.df## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> 28 4.85 1.97
## # ℹ 11 more rows# Subset for turtles that weigh greater than or equal to 10g
subset.turtles.df <- turtles.df %>%
filter(weight >= 10)
subset.turtles.df## # A tibble: 4 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <chr> <dbl> <dbl> <dbl>
## 1 11 female 46.4 8.18 11
## 2 5 female 45 8.05 10.9
## 3 22 female 48.1 8.55 12.8
## 4 7 female 48 8.67 13.5# Subset for only females
fem.turtles.df = turtles.df %>%
filter(sex=="female")
# Here we want to know the mean weight of all females
mean(fem.turtles.df$weight)## [1] 10.02857# or we can summarize the mean weight for males and females at the same time using the following tidyverse syntax ...
turtles.df %>%
group_by(sex) %>%
summarize(meanwt = mean(weight))## # A tibble: 4 × 2
## sex meanwt
## <chr> <dbl>
## 1 fem 6.2
## 2 female 10.0
## 3 male 7.29
## 4 <NA> 2.35# OOPS, looks like we just caught an error!Fixing data using subsetting
Let’s fix the error we just noticed in the way sex was represented in the turtle dataset.
unique(turtles.df$sex) # note the two ways of representing females...## [1] "male" "female" NA "fem"turtles.df$sex[turtles.df$sex=="fem"] <- "female" # correct the error
# or alternatively
turtles.df = turtles.df %>%
mutate(
sex= fct_collapse(sex,
female=c("fem","female"),
male=c("male")))
# or alternatively
turtles.df = turtles.df %>%
mutate(sex = replace(sex,sex=="fem","female"))
turtles.df %>% # summarize weight by sex (check that it's fixed)
group_by(sex) %>%
summarize(meanwt = mean(weight))## # A tibble: 3 × 2
## sex meanwt
## <fct> <dbl>
## 1 female 9.55
## 2 male 7.29
## 3 <NA> 2.35Selecting and renaming columns
Sometimes we only want to work with a small subset of columns from a larger dataset. Or we have column names that are not informative or don’t make sense to us.
# select only the x y and z columns from the diamonds dataset
diamonds2 <- diamonds %>%
select(x,y,z)
# in base R (non tidyverse) you can do this:
diamonds2 <- diamonds[,c("x","y","z")]
# to change the column names, use the "names()" function
names(diamonds2) # extract the column names## [1] "x" "y" "z"names(diamonds2) <- c("length", "width","depth")
# or rename specific variables using 'dplyr' from the 'tidyverse;
diamonds2 <- rename(diamonds2, len = length)Sorting/ordering data
sorting is another common data operation, which helps to visualize and organize data. In R, sorting is typically accomplished using the “order()” function:
*order()** returns the indices of the original (unsorted) vector in the order that they would appear if properly sorted (increasing, by default) – i.e., “10,3,22” becomes “2,1,3” (i.e., to sort this vector in increasing order, you would need to take the second element, then the first, and then the third).
In the ‘tidyverse’ we can use the “arrange” verb to do this!
# Sorting
# The 'order' function returns the indices of the original (unsorted) vector in the order that would sort the vector from lowest to highest...
order(turtles.df$carapace_length)## [1] 3 10 4 20 5 12 13 14 7 11 1 15 6 18 9 17 21 8 2 19 16# To sort a data frame by one vector, you can use "order()"
turtles.df[order(turtles.df$tag_number),]## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 1 <NA> 29.2 5.1 2.38
## 2 2 <NA> 24.3 4.42 1.65
## 3 3 female 42.8 7.32 8.6
## 4 4 male 40 6.6 6.5
## 5 5 female 45 8.05 10.9
## 6 6 female 40 6.53 6.2
## 7 7 female 48 8.67 13.5
## 8 8 <NA> 32 5.35 2.9
## 9 9 male 35 5.74 3.9
## 10 10 male 41 7.15 7.6
## # ℹ 11 more rows# And in decreasing order:
turtles.df[order(turtles.df$tag_number,decreasing=T),]## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 105 male 44 7.1 9
## 2 104 male 44 7.35 9
## 3 22 female 48.1 8.55 12.8
## 4 19 male 42.3 6.77 7.8
## 5 17 female 35.1 6.04 4.5
## 6 16 <NA> 32 5.37 3
## 7 15 <NA> 28.7 4.89 2.18
## 8 14 male 43 6.6 7.2
## 9 13 <NA> 28 4.85 1.97
## 10 12 female 44 7.55 8.9
## # ℹ 11 more rows# or we can use the "arrange" verb in the 'tidyverse':
turtles.df %>%
arrange(carapace_length)## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 2 <NA> 24.3 4.42 1.65
## 2 13 <NA> 28 4.85 1.97
## 3 15 <NA> 28.7 4.89 2.18
## 4 1 <NA> 29.2 5.1 2.38
## 5 16 <NA> 32 5.37 3
## 6 8 <NA> 32 5.35 2.9
## 7 9 male 35 5.74 3.9
## 8 17 female 35.1 6.04 4.5
## 9 4 male 40 6.6 6.5
## 10 6 female 40 6.53 6.2
## # ℹ 11 more rows# or if we want in descending order...
turtles.df %>%
arrange(desc(carapace_length))## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 22 female 48.1 8.55 12.8
## 2 7 female 48 8.67 13.5
## 3 11 female 46.4 8.18 11
## 4 5 female 45 8.05 10.9
## 5 12 female 44 7.55 8.9
## 6 105 male 44 7.1 9
## 7 104 male 44 7.35 9
## 8 14 male 43 6.6 7.2
## 9 3 female 42.8 7.32 8.6
## 10 19 male 42.3 6.77 7.8
## # ℹ 11 more rows# Sorting by 2 columns
turtles.df %>%
arrange(sex,weight)## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 17 female 35.1 6.04 4.5
## 2 6 female 40 6.53 6.2
## 3 3 female 42.8 7.32 8.6
## 4 12 female 44 7.55 8.9
## 5 5 female 45 8.05 10.9
## 6 11 female 46.4 8.18 11
## 7 22 female 48.1 8.55 12.8
## 8 7 female 48 8.67 13.5
## 9 9 male 35 5.74 3.9
## 10 4 male 40 6.6 6.5
## # ℹ 11 more rowsChallenge exercises
Save the “comm_data.txt” file to your working directory. Read this file in as a data frame. Select only the following columns: Hab_class, C_DWN, C_UPS (discard the remaining columns). Finally, rename these columns as: “Class”,“Downstream”, and “Upstream” respectively. [hint 1: use read_table to read in the file as a data frame] [hint 2: use the “select” verb to select the columns you want] [hint 3: use the names() function to rename the columns]
Read in the file “turtle_data.txt”. Create a new version of this data frame with all missing data removed (discard all rows with one or more missing data). Save this new data frame to your project directory as a comma delimited text file. [hint 1: use the “na.omit()” function to remove rows with NAs] [hint 2: use the write_csv function to write to your working directory]
Read in the file “turtle_data.txt”. Create a new data frame with only male turtles. Use this subsetted data set to compute the mean and standard deviation for carapace length of male turtles.
# CHALLENGE EXERCISES -------------------------------------
# 1: Save the "comm_data.txt" file to your working directory. Read this file in as a data frame. Select only only the following columns: Hab_class, C_DWN, C_UPS (discard the remaining columns). Finally, rename these columns as: "Class","Downstream", and "Upstream" respectively. [hint 1: use read_table to read in the file as a data frame] [hint 2: use the "select" verb to select the columns you want] [hint 3: use the names() function to rename the columns]
#
# 2: Read in the file "turtle_data.txt". Create a new version of this data frame with all missing data removed (discard all rows with one or more missing data). Save this new data frame to your project directory as a comma delimited text file. [hint 1: use the "na.omit()" function to remove rows with NAs] [hint 2: use the write_csv function to write to your working directory]
#
# 3: Read in the file "turtle_data.txt". Create a new data frame with only male turtles. Use this subsetted data set to compute the mean and standard deviation for carapace length of male turtles.Bonus: dealing with missing data
In many real-world datasets, observations are incomplete in some way- they are missing information.
In R, the code “NA” (not available) stands in for elements of a vector that are missing for whatever reason.
Most statistical functions have ways of dealing with NAs.
Let’s explore a data set with missing data.
Download this Data with missing values and save to your working directory.
Now let’s explore this data set in more detail:
# Missing Data
# NOTE: you need to specify that this is a tab-delimited file. It is especially important to specify the delimiter for data files with missing data. If you specify the header and what the text is delimited by correctly, it will read missing data as NA. Otherwise it will fail to read data in properly.
missing.df <- read_delim(file="data_missing.txt",delim="\t") # try replacing with "read_table"- it does not work right!
# Missing data are read as an NA
missing.df## # A tibble: 20 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep NA 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 NA A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama NA 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 NA N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela NA 7 N# Can summarize your data and tell you how many NA's per col
summary(missing.df)## Country Import Export Product
## Length:20 Min. :35.00 Min. : 0.00 Length:20
## Class :character 1st Qu.:60.00 1st Qu.: 3.25 Class :character
## Mode :character Median :74.00 Median :11.00 Mode :character
## Mean :70.53 Mean :10.22
## 3rd Qu.:84.00 3rd Qu.:15.75
## Max. :89.00 Max. :23.00
## NA's :3 NA's :2# Omits (removes) rows with missing data
na.omit(missing.df)## # A tibble: 15 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 Ecuador 70 6 A
## 8 ElSalvador 60 13 A
## 9 Guatemala 55 9 A
## 10 Haiti 35 3 A
## 11 Jamaica 87 23 A
## 12 Mexico 83 4 N
## 13 Nicaragua 68 0 A
## 14 Paraguay 74 3 A
## 15 TrinidadTobago 84 15 A# ?is.na (Boolean test!)
is.na(missing.df)## Country Import Export Product
## [1,] FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE FALSE
## [3,] FALSE FALSE FALSE FALSE
## [4,] FALSE FALSE FALSE FALSE
## [5,] FALSE FALSE FALSE FALSE
## [6,] FALSE FALSE FALSE FALSE
## [7,] FALSE TRUE FALSE FALSE
## [8,] FALSE FALSE FALSE FALSE
## [9,] FALSE FALSE FALSE FALSE
## [10,] FALSE FALSE FALSE FALSE
## [11,] FALSE FALSE FALSE FALSE
## [12,] FALSE FALSE TRUE FALSE
## [13,] FALSE FALSE FALSE FALSE
## [14,] FALSE FALSE FALSE FALSE
## [15,] FALSE FALSE FALSE FALSE
## [16,] FALSE TRUE FALSE FALSE
## [17,] FALSE FALSE FALSE FALSE
## [18,] FALSE FALSE TRUE FALSE
## [19,] FALSE FALSE FALSE FALSE
## [20,] FALSE TRUE FALSE FALSEcomplete.cases(missing.df) # Boolean: for each row, tests if there are no NA values## [1] TRUE TRUE TRUE TRUE TRUE TRUE FALSE TRUE TRUE TRUE TRUE FALSE
## [13] TRUE TRUE TRUE FALSE TRUE FALSE TRUE FALSE# Replace all missing values in the data frame with a an interpolation function from tidyverse: replace_na
missing.df %>%
mutate(Export = replace_na(Export,mean(Export,na.rm=T)),
Import = replace_na(Import,mean(Import,na.rm=T)))## # A tibble: 20 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 70.5 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 10.2 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 70.5 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 10.2 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 70.5 7 N# or using tidyverse trickery (less code repetition)
missing.df %>%
mutate(across(where(is.numeric),~replace_na(.,mean(.,na.rm=T))))## # A tibble: 20 × 4
## Country Import Export Product
## <chr> <dbl> <dbl> <chr>
## 1 Bolivia 46 0 N
## 2 Brazil 74 0 N
## 3 Chile 89 16 N
## 4 Colombia 77 16 A
## 5 CostaRica 84 21 A
## 6 Cuba 89 15 A
## 7 DominicanRep 70.5 14 A
## 8 Ecuador 70 6 A
## 9 ElSalvador 60 13 A
## 10 Guatemala 55 9 A
## 11 Haiti 35 3 A
## 12 Honduras 51 10.2 A
## 13 Jamaica 87 23 A
## 14 Mexico 83 4 N
## 15 Nicaragua 68 0 A
## 16 Panama 70.5 19 N
## 17 Paraguay 74 3 A
## 18 Peru 73 10.2 N
## 19 TrinidadTobago 84 15 A
## 20 Venezuela 70.5 7 NAside: the pipe operator (%>%)
The ‘tidyverse’ set of packages takes advantage of the pipe operator
%>%, which provides a clean and intuitive way to
structure code and perform sequential operations in R.

Key advantages include:
code is more readable and intuitive – reads left to right, rather than inside out as is the case for nested function
perform multiple operations without creating a bunch of intermediate (temporary) datasets
This operator comes from the magrittr package, which is included in the installation of all of the tidyverse packages. The shortcut for the pipe operator is ctrl-shift-m (‘m’ is for Magritte). When reading code out loud, use ‘then’ for the pipe. For example the command here:
x %>% log() %>% round(digits=2)
can be interpreted as follows:
Take “x”, THEN take its natural logarithm, THEN round the resulting value to 2 decimal places
The structure is simple. Start with the object you want to manipulate, and apply actions (e.g., functions) to that object in the order in which you want to apply them.
Here is a quick example.
# ASIDE: using the pipe operator %>% (ctrl-shift-m) in R
# start with a simple example
x <- 3
# calculate the log of x
log(x) # form f(x) is equivalent to## [1] 1.098612x %>% log() # form x %>% f## [1] 1.098612# example of multiple steps in pipe
round(log(x), digits=2) # form g(f(x)) is equivalent to## [1] 1.1x %>% log() %>% round(digits=2) ## [1] 1.1Some more subsetting practice:
Returning to the turtle example, we can use subsetting operations to correct or alter data:
# Data Manipulation using subsetting
# list of tags we do not trust the data for
bad.tags <- c(13,105)
turtles.df = turtles.df %>%
mutate(
sex = replace(sex,tag_number%in%bad.tags,NA),
carapace_length = replace(carapace_length,tag_number%in%bad.tags,NA),
head_width = replace(head_width,tag_number%in%bad.tags,NA),
weight = replace(weight,tag_number%in%bad.tags,NA)
)
# or... use some more tidyverse helper functions and tricks!
turtles.df = turtles.df %>%
mutate(across(c("sex","carapace_length","head_width","weight"),
~replace(.,tag_number%in%bad.tags,NA)))
# or use the following non-tidyverse syntax... which still seems easier to me!
turtles.df[turtles.df$tag_number%in%bad.tags,c("sex","carapace_length","head_width","weight")] <- NA
turtles.df## # A tibble: 21 × 5
## tag_number sex carapace_length head_width weight
## <dbl> <fct> <dbl> <dbl> <dbl>
## 1 10 male 41 7.15 7.6
## 2 11 female 46.4 8.18 11
## 3 2 <NA> 24.3 4.42 1.65
## 4 15 <NA> 28.7 4.89 2.18
## 5 16 <NA> 32 5.37 3
## 6 3 female 42.8 7.32 8.6
## 7 4 male 40 6.6 6.5
## 8 5 female 45 8.05 10.9
## 9 12 female 44 7.55 8.9
## 10 13 <NA> NA NA NA
## # ℹ 11 more rows # make a new variable "size.class" based on the "weight" variable
turtles.df = turtles.df %>%
mutate(size.class = case_when(
weight < 3 ~ "juvenile",
weight > 6 ~ "adult",
is.na(weight) ~ NA_character_,
.default = "subadult"
))
turtles.df$size.class## [1] "adult" "adult" "juvenile" "juvenile" "subadult" "adult"
## [7] "adult" "adult" "adult" NA "adult" "juvenile"
## [13] "subadult" "subadult" "adult" "adult" NA "adult"
## [19] "adult" "juvenile" "adult"