Understanding Random Intercepts and Slopes in Mixed-Effects Models using lme4 and glmmTMB
Grecia, Joshua and Lawrence
11-04-2025
Background
Multi-level regression, also known as a hierarchical linear model, is a statistical technique used to analyse data with a hierarchical, nested, or clustered structure, such as students within classrooms or employees within companies. It extends standard regression by accounting for the fact that observations from the same group are not independent, allowing for differences in intercepts and slopes across groups. This approach acknowledges the data structure, prevents biased standard errors, and allows examination of effects at different levels. Real-world data often have hierarchical or repeated structures, and ignoring grouping leads to biased estimates and underestimated uncertainty. Mixed models allow for group-specific variation, partial pooling, and better generalization across groups.
Why multi-levels matter
- Data from the same class will perform more similarly than data from
different contexts.
- Children in the same class will perform more similarly than children from different classes, potentially because they have the same teacher and use the same materials.
- People treated in the same clinics should be more similar in their responses than those treated at different clinics, even if they use the same protocols, etc.
- Lack of Independence
- Violates the assumption of errors being independent
- Bias, SEs, CIs, and p-values
Fixed vs Random Effects
Fixed effects represent population-level parameters that estimate specific coefficients applicable to the entire dataset, such as treatment effects or time trends. In contrast, random effects capture group-level variation by modeling variance components that reflect how individual groups, like schools, subjects, or sites, deviate from the overall average. While fixed effects provide estimates of average trends, random effects account for the variability among groups, allowing for partial pooling and more accurate inference.
Nested vs Crossed Random Effects
- Nested: One factor is contained within another Syntax: (1 | school/classroom)
- Crossed: Factors interact independently Syntax: (1 | student) + (1 | teacher)
Benefits of multi-level models
- Modelling variability in effects across contexts (quantify variation
among groups)
- Model the variability in intercepts
- Model the variability in slopes
- Gives more realistic predictions (we don’t force every group to behave identically)
- Reduces bias in estimated effects and standard errors.
Mixed-effects models are used when data are grouped or clustered. For example, students nested within schools, patients measured by different doctors, or repeated measures on individuals over time.
These models allow us to:
- Model population-level effects (fixed effects): e.g., the overall effect of treatment.
- Model group-level variation (random effects): e.g., how the treatment effect or baseline differs across different groups.
These characteristics makes them particularly valuable when we expect differences among groups but want to share information across them.
When to use which?
| Scenario | Recommended Package |
|---|---|
| Simple Gaussian or Poisson GLMM | lme4 |
| Zero-inflated or overdispersed counts | glmmTMB |
| Need for hurdle or beta models | glmmTMB |
| Fast prototyping with standard families | lme4 |
| Diagnostics with simulated residuals | glmmTMB + DHARMa |
We will focus on
lmerto explain random slopes and intercepts
Random Intercepts and Random Slopes
When we fit a mixed-effects model using lmer, we can
include random intercepts and random
slopes.
- Random intercept: Each group has its own baseline outcome.
- Random slope: Each group can differ in how strongly a predictor (like treatment) affects the outcome.
Mathematically:
\[ y_{ij} = (\beta_0 + u_{0j}) + (\beta_1 + u_{1j})x_{ij} + \epsilon_{ij} \]
Where:
- \(\beta_0\), \(\beta_1\) = fixed (overall) intercept and
slope
- \(u_{0j}\), \(u_{1j}\) = random deviations for group
j
- \(\epsilon_{ij}\) = residual error
Syntax in lme4
| Model Type | Formula | Meaning |
|---|---|---|
| Random intercept only | (1 | group) |
Groups have different intercepts |
| Random intercept and slope | (1 + treatment | group) |
Groups have different intercepts and treatment effects |
| Crossed random effects | (1 + treatment | A) + (1 | B) |
Effects vary across two independent grouping factors |
Correlation Between Intercepts and Slopes
When we specify (1 + treatment | group), the model also
estimates how correlated the random intercepts and slopes are.
\[ \rho = \frac{\sigma_{u_0, u_1}}{\sigma_{u_0} \sigma_{u_1}} \]
The estimated correlation can ether be positive or negative.
- A positive correlation means schools with higher
intercepts also tend to have stronger treatment effects.
- A negative correlation means schools with higher intercepts tend to have smaller treatment effects.
Let us work through examples for nested and crossed random effects
We will do the following
- Go through data simulation for running both model types
- Extract and interpret results
- Brace up for a real data. It’s going to be fun
Workflow 1: Nested Random Effects (Schools and Students)
- We’ll simulate a dataset where students are nested within schools, and each school has its own baseline and treatment effect. Look at this as students inside school.
- We are working with two hypothetical treatments namely control and treated.
- control and treated can be interpreted as two distinct sets of students (e.g., male vs female; undergraduate vs graduate, etc)
knitr::opts_chunk$set(echo = TRUE)library(lme4)## Loading required package: Matrixlibrary(glmmTMB)
library(tidyverse)## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.5
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 4.0.0 ✔ tibble 3.3.0
## ✔ lubridate 1.9.4 ✔ tidyr 1.3.1
## ✔ purrr 1.1.0## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ tidyr::expand() masks Matrix::expand()
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ✖ tidyr::pack() masks Matrix::pack()
## ✖ tidyr::unpack() masks Matrix::unpack()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(ggplot2)
library(dplyr)
library(broom.mixed)1.1 Data Generation for Nested Model
set.seed(123)
n_schools <- 5
n_students <- 50
df_nested <- data.frame(
school = factor(rep(1:n_schools, each = n_students)),
treatment = factor(rep(rep(c("control","treated"), each = n_students/2), times = n_schools))
)
# Random intercepts and slopes per school
school_intercepts <- rnorm(n_schools, 0, 1)
school_slopes <- rnorm(n_schools, 2, 0.5)
df_nested$y <- 10 +
school_intercepts[as.numeric(df_nested$school)] +
school_slopes[as.numeric(df_nested$school)] * (df_nested$treatment == "treated") +
rnorm(nrow(df_nested), 0, 1)
head(df_nested)## school treatment y
## 1 1 control 10.663606
## 2 1 control 9.799338
## 3 1 control 9.840296
## 4 1 control 9.550207
## 5 1 control 8.883683
## 6 1 control 11.226437str(df_nested)## 'data.frame': 250 obs. of 3 variables:
## $ school : Factor w/ 5 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ treatment: Factor w/ 2 levels "control","treated": 1 1 1 1 1 1 1 1 1 1 ...
## $ y : num 10.66 9.8 9.84 9.55 8.88 ...y = Response variable
treatment = Predictor variable
school = Grouping variable
1.2 Fitting the Nested Model
We will fit a mix effect model using school as the grouping variable and specifying a random intercept and slope for each school.
m_nested <- lmer(y ~ treatment + (1 + treatment | school), data = df_nested)
summary(m_nested)## Linear mixed model fit by REML ['lmerMod']
## Formula: y ~ treatment + (1 + treatment | school)
## Data: df_nested
##
## REML criterion at convergence: 704.8
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.3836 -0.6928 -0.1003 0.6241 3.3547
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## school (Intercept) 0.5040 0.7100
## treatmenttreated 0.3148 0.5611 -0.84
## Residual 0.9054 0.9515
## Number of obs: 250, groups: school, 5
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 10.1510 0.3287 30.881
## treatmenttreated 2.0430 0.2783 7.341
##
## Correlation of Fixed Effects:
## (Intr)
## tretmnttrtd -0.812Interpretation
- Fixed effects: Average (overall) intercept and
treatment effect.
- Random effects: Variance of intercepts and slopes
across schools, plus their correlation.
- Residual variance: Variation not explained by school or treatment.
1.3 Extracting Random Effects
fix <- fixef(m_nested)
ranefs <- ranef(m_nested)$school
school_effects <- data.frame(
school = rownames(ranefs),
intercept = fix["(Intercept)"] + ranefs[,"(Intercept)"],
treatment_effect = fix["treatmenttreated"] + ranefs[,"treatmenttreated"]
)
school_effects## school intercept treatment_effect
## 1 1 9.544692 2.741017
## 2 2 9.728777 2.375214
## 3 3 11.303966 1.449906
## 4 4 10.119412 1.785368
## 5 5 10.058035 1.863547Each row shows how the intercept and treatment effect differ per school. treatment effect is obtained by adding random effects to fixed effect.
1.4 Visualizing Random Slopes
ggplot(school_effects, aes(x = school, y = treatment_effect)) +
geom_point(size = 3) +
geom_hline(yintercept = fix["treatmenttreated"], linetype = "dashed", color = "red") +
labs(y = "Estimated treatment effect", title = "School-specific treatment effects") +
theme_minimal()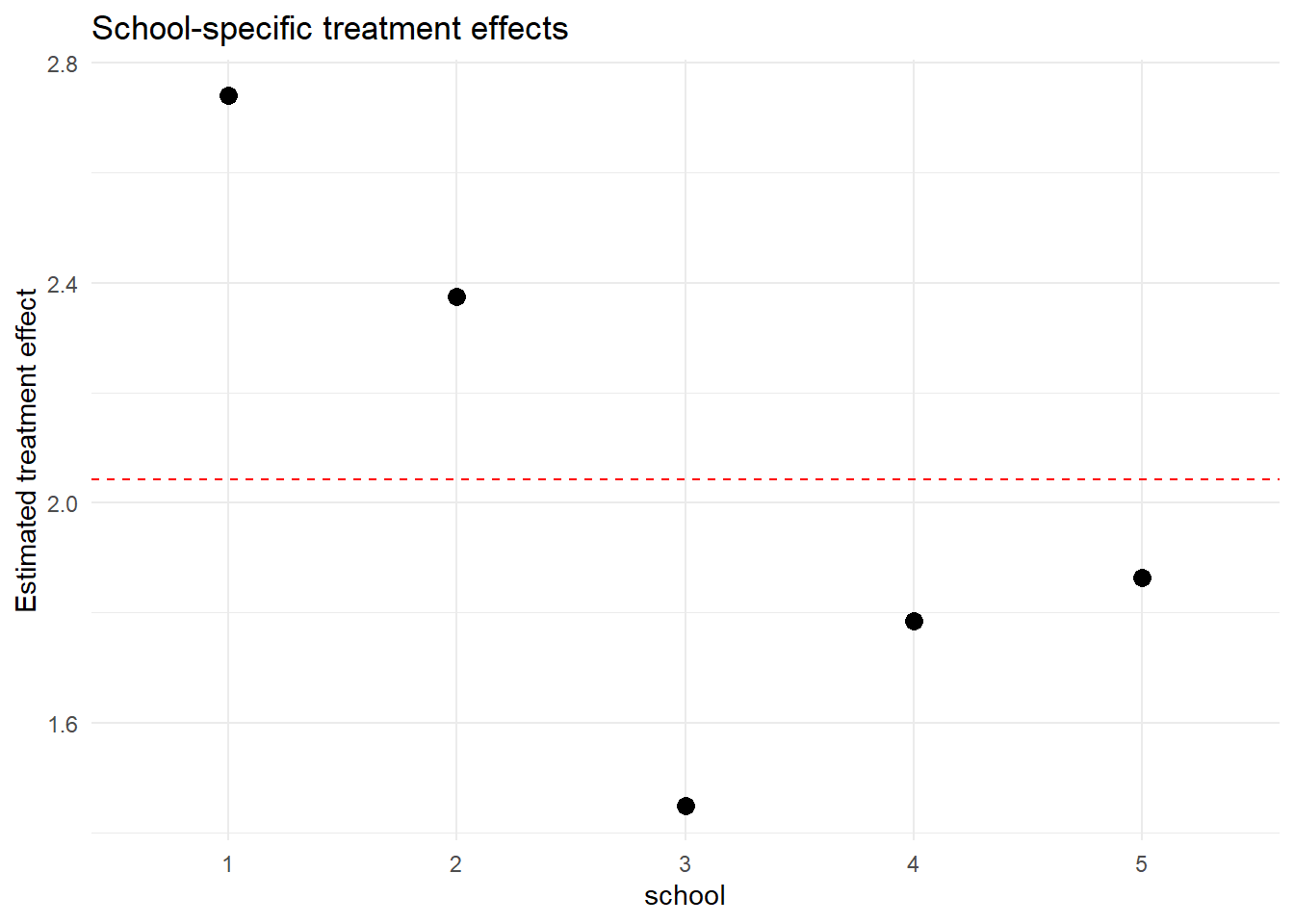
The dashed red line shows the average treatment effect, while the points show each school’s deviation.
1.5 Predicted Lines per School
df_nested$pred <- predict(m_nested)
ggplot(df_nested, aes(x = treatment, y = y, group = school, color = school)) +
stat_summary(fun = mean, geom = "point", size = 3, position = position_dodge(0.3)) +
stat_summary(fun = mean, geom = "line", aes(group = school), size = 1) +
labs(y = "Outcome", title = "Varying treatment effects by school") +
theme_minimal()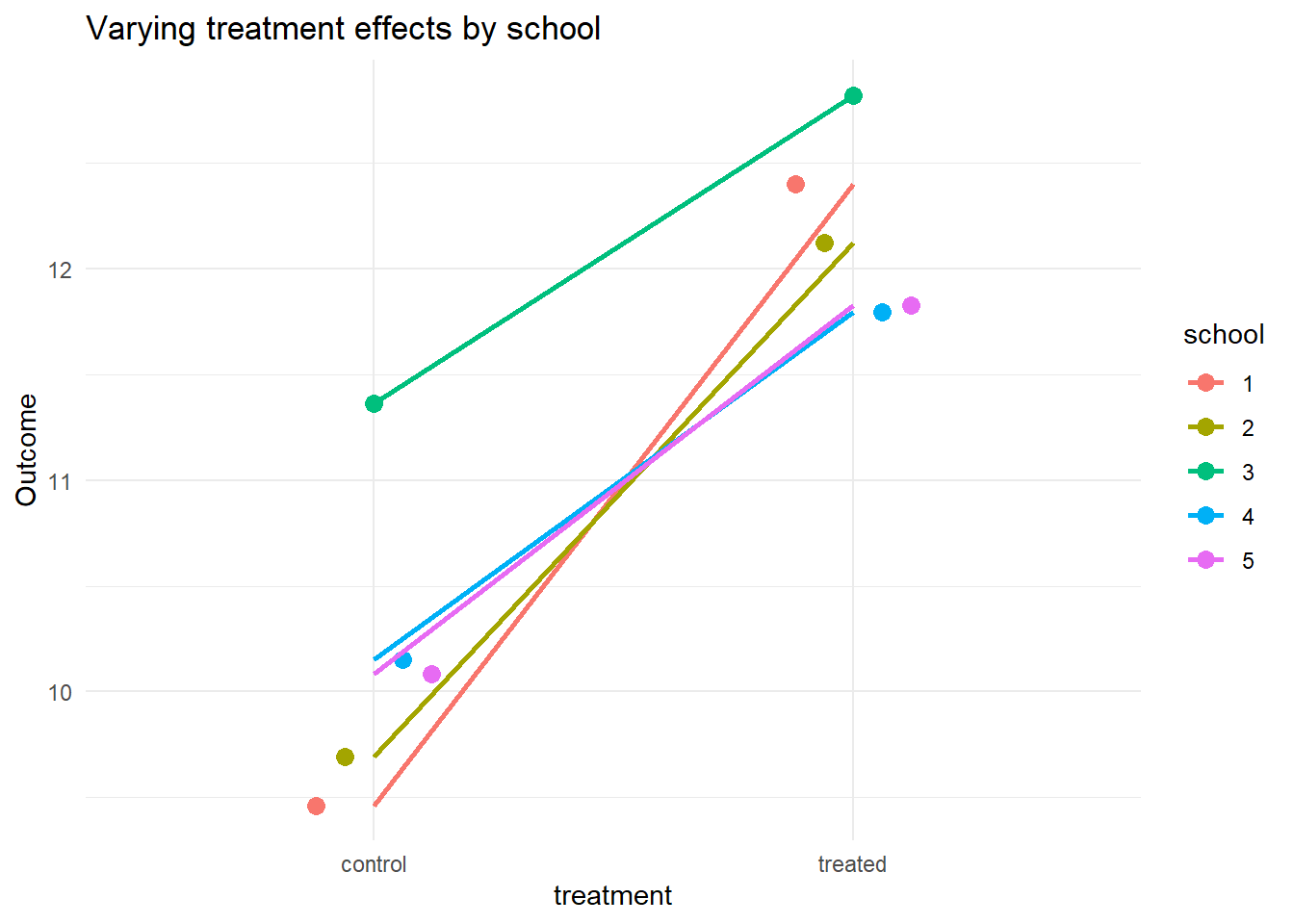
Each line shows how a school responds differently to treatment — some respond more strongly, some less.
1.6 Correlation Between Random Intercepts and Slopes
vc <- VarCorr(m_nested)
vc## Groups Name Std.Dev. Corr
## school (Intercept) 0.70996
## treatmenttreated 0.56111 -0.842
## Residual 0.95155attr(vc$school, "correlation")[1, 2]## [1] -0.8417701This value shows how much the intercepts and slopes co-vary. A positive number means that schools with higher baselines tend to have stronger treatment effects, and vice versa. Here, we have a strong negative correlation, meaning that schools with lower baselines will have stronger treatment effects.
Workflow 2: Crossed Random Effects (Students × Raters)
Now we simulate a crossed structure, specifying that each student is rated by multiple raters. Just like we’ve done, we will:
- Simulate a dataset where each student can be rated by multiple raters.
- We will maintain the two hypothetical treatments namely control and treated.
- Remeber, control and treated can be interpreted as two distinct sets of students (e.g., male vs female; undergraduate vs graduate, etc)
2.1 Data Generation for Crossed Model
set.seed(123)
n_students <- 40
n_raters <- 6
df_crossed <- expand.grid(
student = factor(1:n_students),
rater = factor(1:n_raters),
treatment = factor(c("control","treated"))
)
rater_intercepts <- rnorm(n_raters, 0, 2)
rater_slopes <- rnorm(n_raters, 3, 0.7)
df_crossed$score <- 50 +
rater_intercepts[as.numeric(df_crossed$rater)] +
rater_slopes[as.numeric(df_crossed$rater)] * (df_crossed$treatment == "treated") +
rnorm(nrow(df_crossed), 0, 2)
head(df_crossed)## student rater treatment score
## 1 1 1 control 49.68059
## 2 2 1 control 49.10041
## 3 3 1 control 47.76737
## 4 4 1 control 52.45287
## 5 5 1 control 49.87475
## 6 6 1 control 44.94581str(df_crossed)## 'data.frame': 480 obs. of 4 variables:
## $ student : Factor w/ 40 levels "1","2","3","4",..: 1 2 3 4 5 6 7 8 9 10 ...
## $ rater : Factor w/ 6 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ treatment: Factor w/ 2 levels "control","treated": 1 1 1 1 1 1 1 1 1 1 ...
## $ score : num 49.7 49.1 47.8 52.5 49.9 ...
## - attr(*, "out.attrs")=List of 2
## ..$ dim : Named int [1:3] 40 6 2
## .. ..- attr(*, "names")= chr [1:3] "student" "rater" "treatment"
## ..$ dimnames:List of 3
## .. ..$ student : chr [1:40] "student=1" "student=2" "student=3" "student=4" ...
## .. ..$ rater : chr [1:6] "rater=1" "rater=2" "rater=3" "rater=4" ...
## .. ..$ treatment: chr [1:2] "treatment=control" "treatment=treated"score = Response variable
treatment = Predictor variable as before
student = Grouping variable A
rater = Grouping variable B
2.2 Fitting the Crossed Model
We will specify tw independent grouping factors, necessitate a crossed model structure
m_crossed <- lmer(score ~ treatment + (1 + treatment | rater) + (1 | student), data = df_crossed)
summary(m_crossed)## Linear mixed model fit by REML ['lmerMod']
## Formula: score ~ treatment + (1 + treatment | rater) + (1 | student)
## Data: df_crossed
##
## REML criterion at convergence: 2035.6
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9650 -0.6181 -0.0653 0.6521 3.3913
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## student (Intercept) 0.02136 0.1462
## rater (Intercept) 3.09372 1.7589
## treatmenttreated 0.48899 0.6993 0.28
## Residual 3.80187 1.9498
## Number of obs: 480, groups: student, 40; rater, 6
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 50.8492 0.7294 69.716
## treatmenttreated 3.1800 0.3364 9.453
##
## Correlation of Fixed Effects:
## (Intr)
## tretmnttrtd 0.167Interpretation
- Random intercepts/slopes for raters mean each rater interprets the treatment differently.
- Random intercept for students adjusts for repeated measures per student.
2.3 Extracting Rater Effects
fix <- fixef(m_crossed)
ranefs <- ranef(m_crossed)$rater
rater_effects <- data.frame(
rater = rownames(ranefs),
intercept = fix["(Intercept)"] + ranefs[,"(Intercept)"],
treatment_effect = fix["treatmenttreated"] + ranefs[,"treatmenttreated"]
)
rater_effects## rater intercept treatment_effect
## 1 1 49.03759 3.503371
## 2 2 49.64948 2.447363
## 3 3 52.77535 2.944621
## 4 4 49.94620 2.614150
## 5 5 50.45341 3.621322
## 6 6 53.23342 3.9494712.4 Visualizing Rater Treatment Effects
ggplot(rater_effects, aes(x = rater, y = treatment_effect)) +
geom_point(size = 3) +
geom_hline(yintercept = fix["treatmenttreated"], linetype = "dashed", color = "red") +
labs(y = "Estimated treatment effect", title = "Rater-specific treatment effects") +
theme_minimal()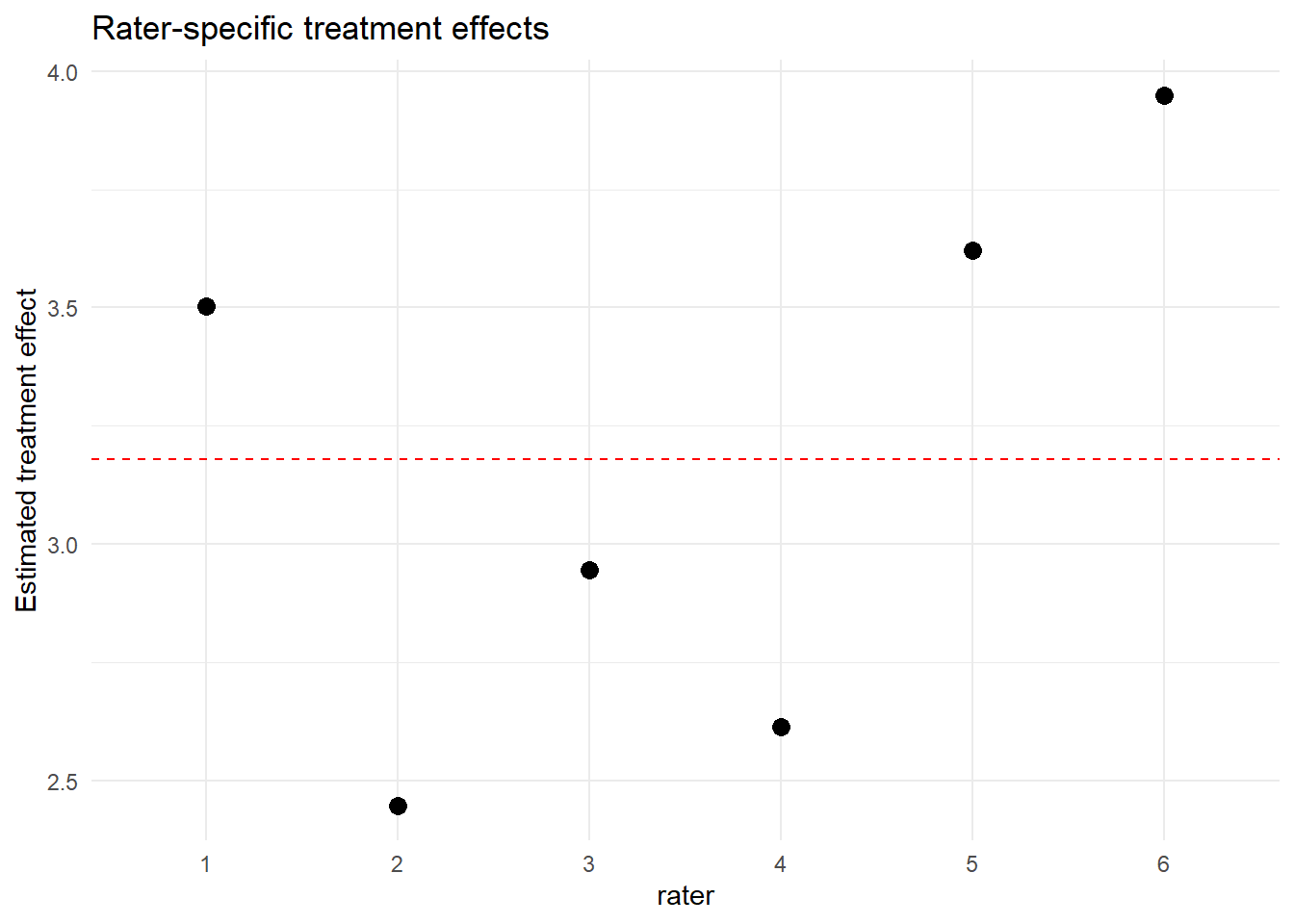
2.5 Predicted Scores by Rater
df_crossed$pred <- predict(m_crossed)
ggplot(df_crossed, aes(x = treatment, y = pred, group = rater, color = rater)) +
stat_summary(fun = mean, geom = "point", size = 3, position = position_dodge(0.3)) +
stat_summary(fun = mean, geom = "line", aes(group = rater), size = 1) +
labs(y = "Predicted score", title = "Varying treatment effects by rater") +
theme_minimal()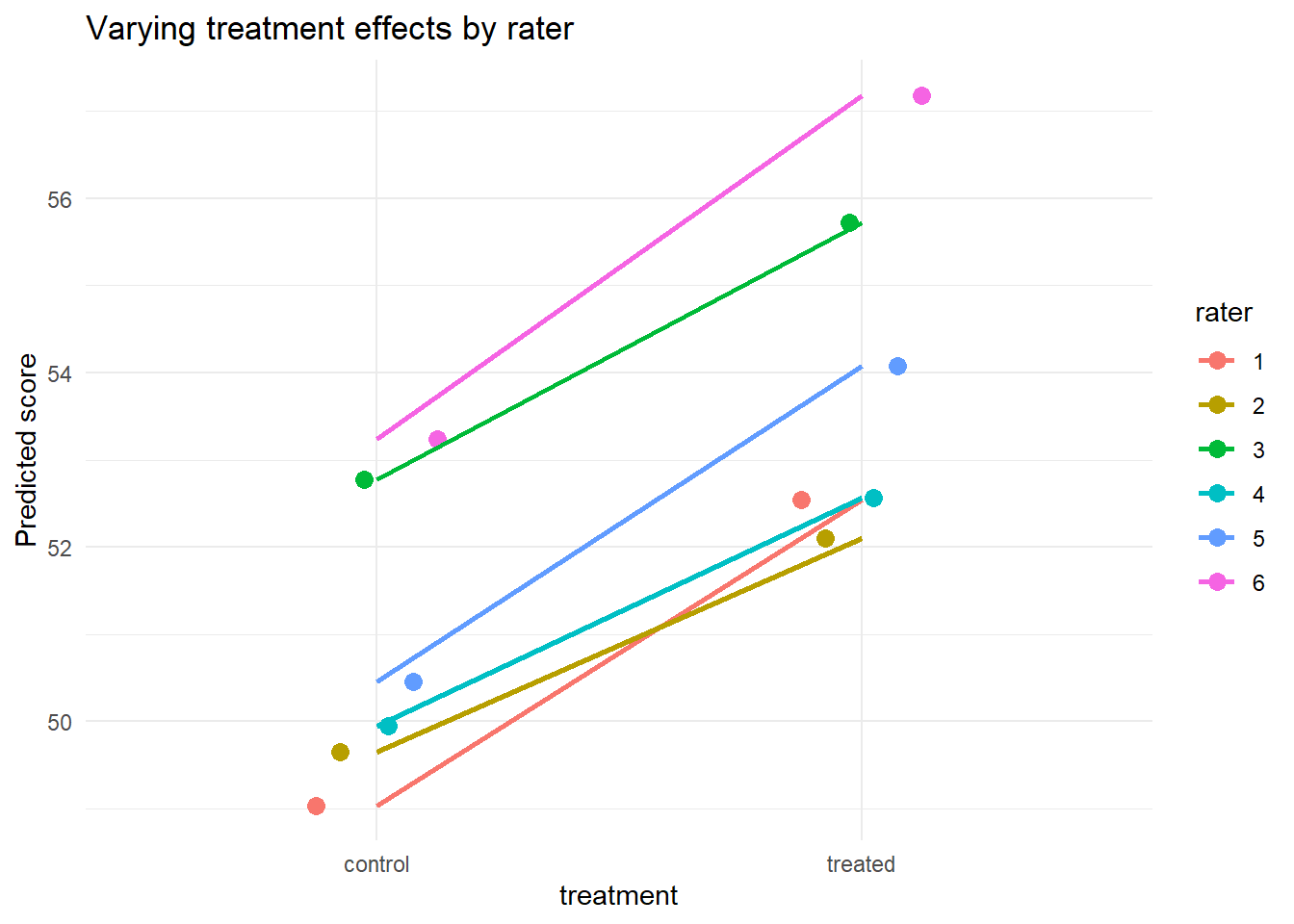
Each rater interprets “treatment” differently, leading to variability in slopes.
2.6 Random Effect Correlation
VarCorr(m_crossed)## Groups Name Std.Dev. Corr
## student (Intercept) 0.14617
## rater (Intercept) 1.75890
## treatmenttreated 0.69928 0.277
## Residual 1.94984The covariance section again shows how intercepts and slopes for raters co-vary.Here, there is a weak positive correlation, meaning that raters with higher baselines have stronger treatment effects.
Summary
Let’s do a quick recap:
- Random intercepts capture baseline variability
across groups.
- Random slopes capture variability in the effect
of a predictor across groups.
- Correlation between intercepts and slopes tells whether groups with
higher baselines show stronger or weaker responses.
- Mixed models balance flexibility (allowing variation) with efficiency (borrowing strength across groups).
Hands-On Exercise
Let’s do a quick class work to demonstrate what we have learnt so far. Here, we are going to use a real dataset present in the lme4 package.
Objective:
Fit and interpret a mixed-effects model with random slopes and intercepts using a new dataset.
Dataset:
Use the built-in sleepstudy dataset in
lme4. Run ?sleepstudy to understand that
dataset.
data("sleepstudy")
head(sleepstudy)## Reaction Days Subject
## 1 249.5600 0 308
## 2 258.7047 1 308
## 3 250.8006 2 308
## 4 321.4398 3 308
## 5 356.8519 4 308
## 6 414.6901 5 308Tasks:
Fit a model predicting
Reactionas a function ofDayswith random intercepts and slopes forSubject:m_exercise <- lmer(Reaction ~ Days + (1 + Days | Subject), data = sleepstudy) summary(m_exercise)Interpret:
- What do the fixed effects mean?
- How do random intercepts and slopes vary across subjects?
- What does the correlation between them imply?
Plot subject-specific slopes using:
coef(m_exercise)$SubjectDiscuss:
- Why do some subjects show stronger or weaker effects of
DaysonReaction?
- What happens if we remove the random slope term?
- Why do some subjects show stronger or weaker effects of