Small-population paradigm
NRES 470/670
Spring 2024
The final extinction of a population is usually caused by factors that small populations are uniquely sensitive to; once a population gets small enough, demographic stochasticity, genetic drift, and inbreeding depression can together deliver the ‘final blow’ to a population.
In conservation biology the study of the drivers of extinction for small populations is called the small-population paradigm. The small-population paradigm refers to the tendency in conservation biology to study those largely stochastic factors that can result in the extinction or degradation of small populations.
Demographic stochasticity
We have already explored this concept, which is central to the small population paradigm!
The simple fact is: it is much more likely for all 10 individuals in a population to be unlucky in a given year than for all 1000 individuals in a population to be unlucky in a given year!! – just as it is very unlikely for all coin flips out of 1000 to come up heads…
This illustrates the very important concept that small populations can go extinct due to demographic stochasticity alone, whereas this possibility is vanishingly small for large populations.
The moral of the story is: weird and dramatic things (e.g., extinction) can happen in small populations for no other reason than that the population is small!
Dramatic events like random extinction just would never happen in large populations due to random mating, birth, and mortality processes (assuming a stable, favorable environment and no range-wide catastrophic events)!
Genetic drift
Same goes for genetic drift! Severe or consequential random fluctuations in a population’s gene pool (genetic drift) is a small-population phenomenon!
In large populations, random forces are just not strong enough to eradicate a relatively common gene variant (allele) from a population (this could happen by natural selection, but not through random reproductive processes).
In small populations, random chance can be the primary driver of genetic change over time!
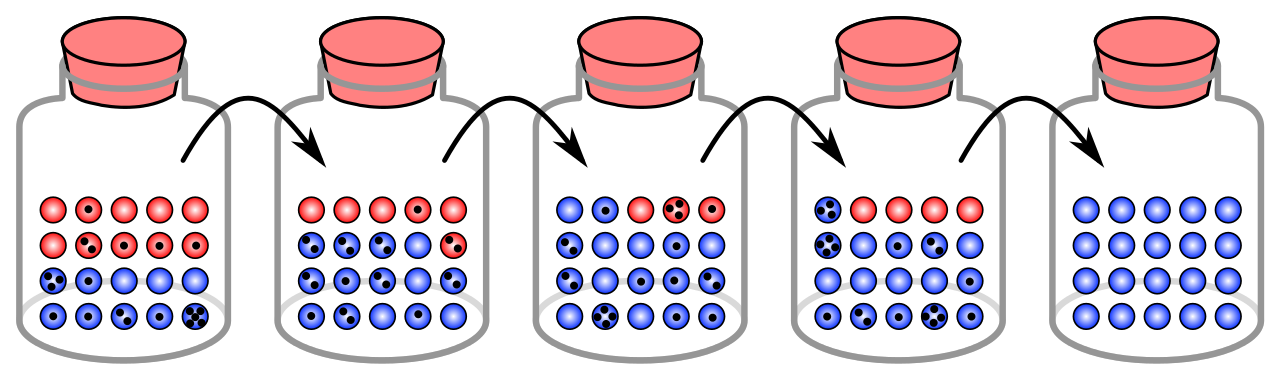
Let’s imagine that the balls in the jars in the above figure are individuals. The black dots indicate how many offspring each individual will have (thereby passing on its genes). The different colored balls represent different genetic variants. Each jar, left to right, represents a different (non-overlapping) generation.
Does this sound familiar? Just by random chance, some individuals will breed and some will not. Just by random chance, some individuals will have more of their offspring survive. Genetic drift is a consequence of demographic stochasticity.
Finally, let’s imagine that the red balls represent individuals that are more resilient to drought (but have very little selective advantage under normal circumstances). Just by random chance, the blue individuals happen to successfully mate and reproduce more than the red individuals- the blue variant becomes fixed in the population and the red variants go extinct. There is no natural selection involved in this scenario – just stochasticity!
And now, by random chance, this population is now more susceptible to drought than it was before!
In a much larger population, the possibility of losing this drought-resistance allele from the gene pool due to random genetic drift would be vanishingly small.
Inbreeding depression
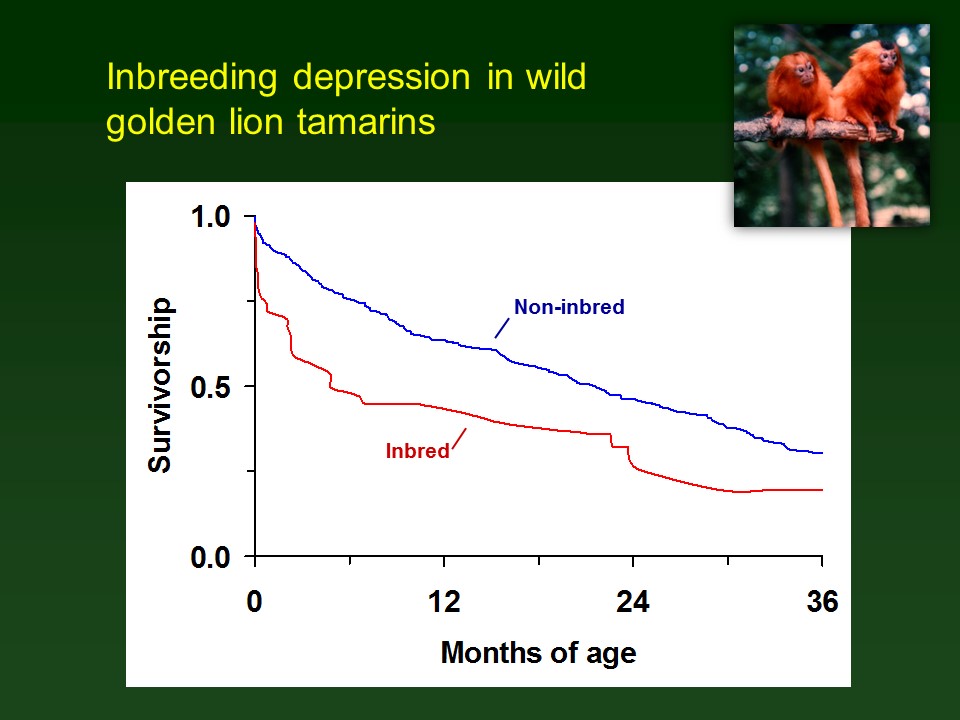
Inbreeding depression is another problem peculiar to small populations.
Most diploid populations have some ‘bad’ gene variants (alleles) in their gene pool. These are often called deleterious alleles. These deleterious alleles usually don’t cause problems because in general they are recessive - that is, the bad effect is only apparent if an individual has two “bad” copies of that allele! If these alleles are rare in the population, it is extremely unlikely that you get two copies of the same bad allele by chance!
Unless… the two parents are close relatives (which can be more common in small populations). If parents are close relatives, both parents have a relatively high probability of inheriting at least one of the same ‘bad’ gene variants from a single common ancestor!
If you consider that that single common ancestor likely had deleterious recessive alleles embedded in their genome at many unique loci, then the chance of an inbred offspring inheriting two deleterious copies in at least one of those loci becomes fairly high!
In a way, inbreeding depression and genetic drift represents a kind of Allee effect, where population vital rates can deteriorate as the population size is reduced (positive density dependence). Unfortunately, unlike the Allee effects we have studied earlier in this class, this kind of Allee effect can have long-term consequences even if the population size is increased, since the gene pool can remain degraded for many generations after “recovery”.
Extinction vortex
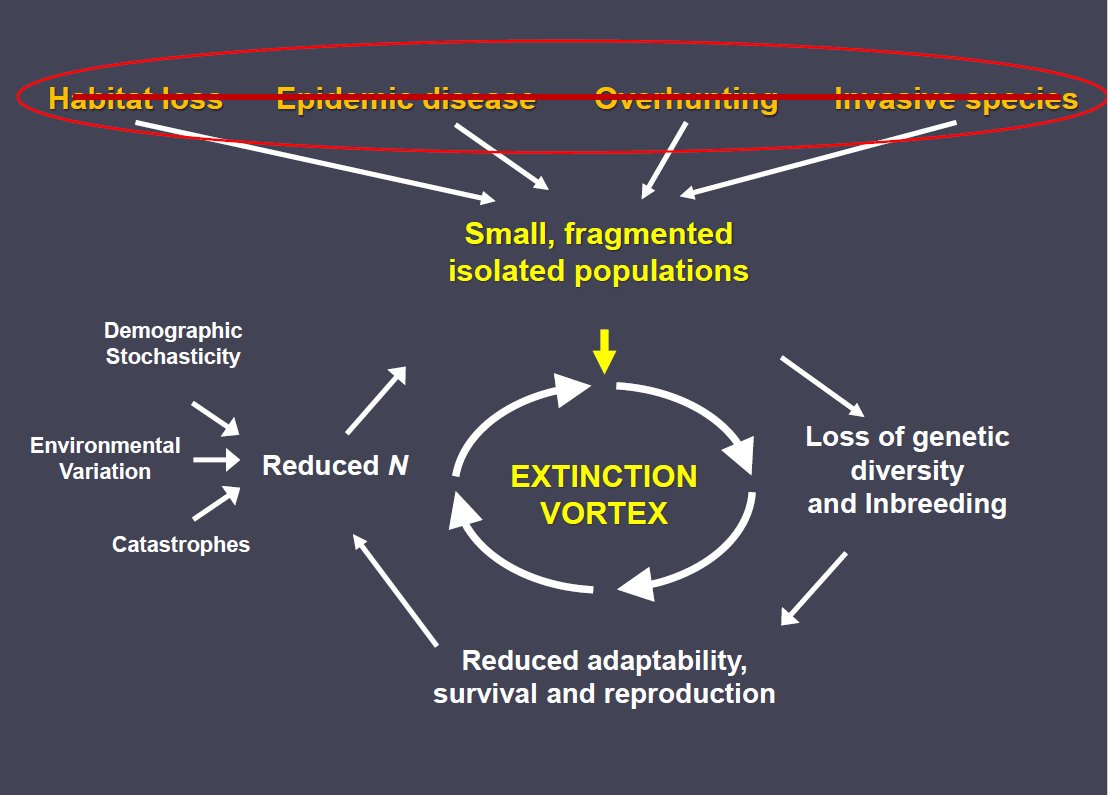
One of the important ideas emerging from the small population paradigm is the concept of the extinction vortex. This concept is illustrated in the above figure.
In general, large and functional populations become small as a result of deterministic “threat” factors like harvest and habitat loss (this is the topic of the next lecture on the “declining population paradigm”)
Once a population gets small enough, it’s range generally contracts as well, making it more vulnerable to environmental stochasticity- including catastrophic events like fires and floods.
Very small populations also can experience random declines due to demographic stochasticity. Demographic stochasticity can also lead to random loss of useful genes (genetic drift), leading to reduced vital rates and/or reduced adaptive potential.
Small populations are also subject to negative effects of inbreeding depression.
All of this can lead to smaller population sizes, which can leading to further genetic degradation, leading to greater risks from purely stochastic forces like demographic stochasticity. This “vortex” ultimately ends in extinction…
Furthermore, once a population falls into the extinction vortex, it can’t necessarily be saved by removing the factors that caused the population to become small in the first place. That is, once genetic drift has removed important adaptive variation from the gene pool, and once inbreeding has caused deleterious recessive alleles to start expressing more frequently, the population may not be able to recover effectively.
Case study: Florida panther genetic rescue.

In general, extinction is often caused by stochastic processes, but stochastic factors are unlikely to be the reason the population got small in the first place! (that is the topic of the ‘declining population paradigm’ lecture!)
Minimum Viable Populations (MVP)
As wildlife managers, we often wish to know: “how small does a population have to get before we have to start worrying about adverse consequences like extinction, genetic erosion, and inbreeding?”
The most satisfying general definition of MVP (in my opinion) directly relates to the extinction vortex concept: Minimum Viable Population size (MVP) is that population size below which the population becomes vulnerable to the extinction vortex (or alternatively, the abundance threshold above which the extinction vortex can be safely avoided!).
However, the above definition is not strictly quantifiable for most species, since inbreeding depression can be difficult to predict. Also MVP is often defined in relation to population models (like the ones we build in InsightMaker), many of which don’t have an explicit genetic component. The quantitative, operational definition that we will use in this course is:
MVP: the abundance threshold below which extinction risk exceeds [a risk tolerance threshold] over [a time horizon]
To quantify MVP, we need to specify: (1) a time horizon (e.g., 50 years) and (2) a risk tolerance threshold (e.g., 5% extinction risk).
For example:
MVP: the abundance threshold below which extinction risk exceeds 5% over 50 years
Population Viability Analysis (PVA)
Population Viability Analysis (PVA) is often used to model the processes involved in the extinction vortex (the central concept of the small population paradigm). This can include genetic drift and inbreeding depression. However, in this course, we won’t get into modeling genetics (unless you really want to- I’m happy to show you how to do this using the Vortex software!)
One of the most widely-used PVA software packages, Vortex, gets its name from the extinction vortex concept. Vortex does allow explicit modeling of inbreeding and loss of genetic diversity in small populations.

Example: Aruba island rattlesnake
The Aruba Island Rattlesnake, or Cascabel (Crotalus durissus unicolor), is the top predator on the island of Aruba, and primarily consumes rodents.

The Aruba island rattlesnake, as you might expect, occurs only on the island of Aruba.
The Aruba rattlesnake was previously listed as Critically Endangered by IUCN (it is no longer listed as of 2021 due to lack of information), and has several attributes that make it particularly susceptible to falling into the extinction vortex:
See Lab 5 for the related MVP activity.
Q Why are island-endemic species like the Aruba rattlesnake so vulnerable to extinction? [Top Hat]
Q Are species that are confined to small areas (like the Aruba rattlesnake) more vulnerable to environmental stochasticity than species that occupy larger geographic ranges? [Top Hat]